import numpy as np
import matplotlib.pyplot as plt
from bisect import bisect_right
from molass.SEC.Models.Simple import gaussian
from molass.LowRank.LowRankInfo import get_denoised_data
def plot_structure_factor(Rg=35, ratio=1.0, error_level=0.05):
x = np.arange(300)
q = np.linspace(0.005, 0.3, 200)
R = np.sqrt(5/3)*Rg
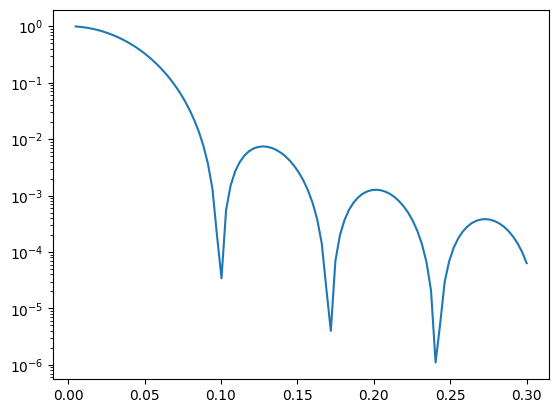
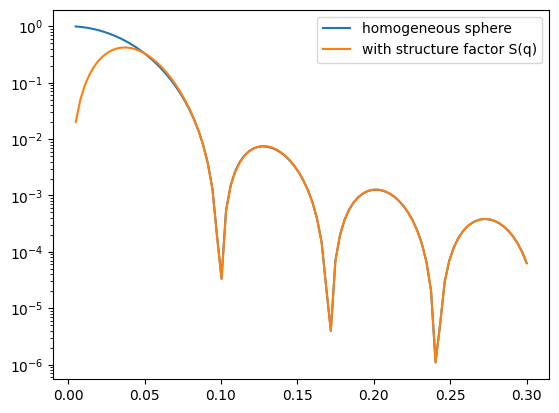
I1 = homogeneous_sphere(q, R)**2
I2 = I1 * S0(q, R)
i = bisect_right(q, 0.02)
h, m, s = 1, 150, 30
y = gaussian(x, h, m, s)
fig = plt.figure(figsize=(15,8))
ax1 = fig.add_subplot(231)
ax2 = fig.add_subplot(232)
ax3 = fig.add_subplot(233, projection='3d')
ax4 = fig.add_subplot(234)
ax5 = fig.add_subplot(235)
ax6 = fig.add_subplot(236, projection='3d')
ax2.set_yscale('log')
for ax in [ax1, ax2]:
ax.plot(q, I1, label='I1: P(q)')
ax.plot(q, I2, label='I2: P(q)S(q)')
bq = (I1 - I2)*ratio
# ax.plot(q, bq, label='(I1 - I2)')
ax.legend()
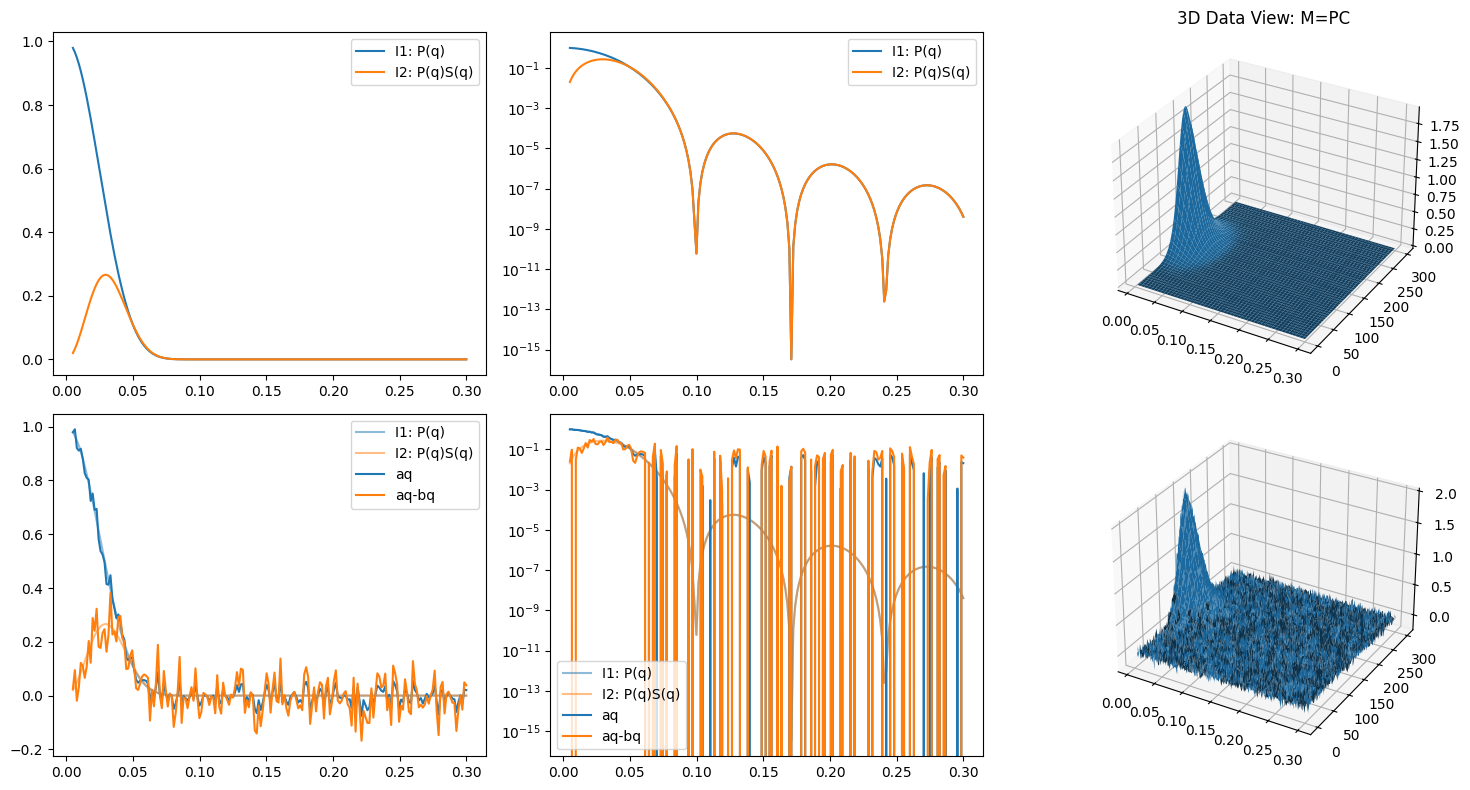
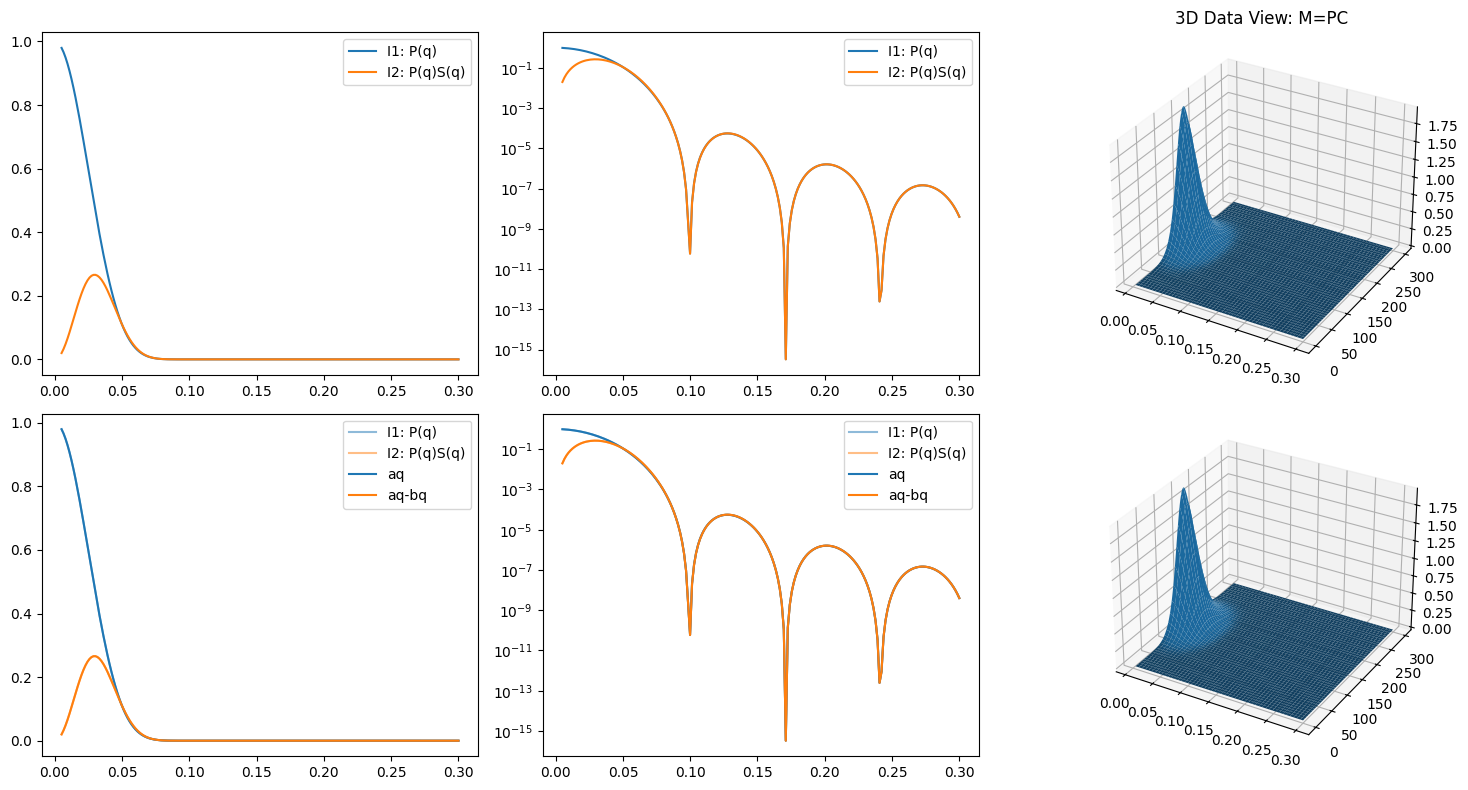
P = np.array([I1, bq]).T
C = np.array([y, y**2])
M = P @ C # matrix multiplication
Me = M + error_level * np.random.randn(*M.shape)
# Me_ = get_denoised_data(Me, rank=2)
# y_ = Me_[i,:]
# Cinv = np.linalg.pinv(np.array([y_, y_**2]))
Cinv = np.linalg.pinv(C)
P_ = Me @ Cinv
xx, qq = np.meshgrid(x, q)
ax3.set_title("3D Data View: M=PC")
ax3.plot_surface(qq, xx, M)
ax6.plot_surface(qq, xx, Me)
ax5.set_yscale('log')
for ax in [ax4, ax5]:
ax.plot(q, I1, alpha=0.5, label='I1: P(q)')
ax.plot(q, I2, alpha=0.5, label='I2: P(q)S(q)')
# ax.plot(q, bq, alpha=0.5, label='(I1 - I2)')
aq = P_[:,0]
bq = P_[:,1]
ax.plot(q, aq, color='C0', label='aq')
# ax.plot(q, bq, color='C2', label='bq')
ax.plot(q, aq - bq, color='C1', label='aq-bq')
ax.legend()
fig.tight_layout()