6.1. Fourier Transform - from Density to Scattering#
Since these explanations are beyond the scope of this book, we take the following relationships, depicted below, for granted:
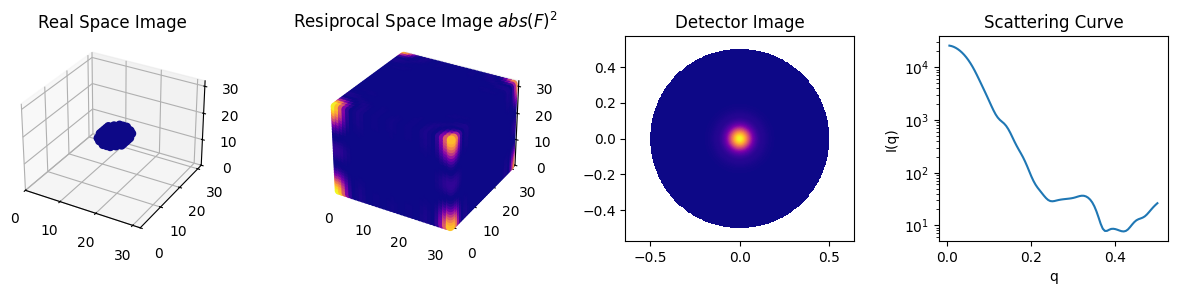
The scattering curve is a circular average of the detector image.
The detector image is a spherical average of the squared absolute values in reciprocal space.
The reciprocal space is obtained by applying a Fourier transform to the real space.
Confirm each relationship in the code.
import numpy as np
import matplotlib.pyplot as plt
from learnsaxs import draw_voxles_as_dots, get_detector_info, draw_detector_image
def plot_an_ellipsoid_with_ft_squared_and_detector_image(N, center, a, b, c):
x = y = z = np.arange(N)
xx, yy, zz = np.meshgrid(x, y, z)
cx, cy, cz = center
shape = (xx - cx)**2/a**2 + (yy - cy)**2/b**2 + (zz - cz)**2/c**2 < 1
canvas = np.zeros((N,N,N))
canvas[shape] = 1 # density inside the ellipsoid is 1, outside 0
fig = plt.figure(figsize=(12,3))
ax1 = fig.add_subplot(141, projection="3d")
ax2 = fig.add_subplot(142, projection="3d")
ax3 = fig.add_subplot(143)
ax4 = fig.add_subplot(144)
ax4.set_yscale("log")
ax1.set_title("Real Space Image")
ax2.set_title("Resiprocal Space Image $abs(F)^2$")
ax3.set_title("Detector Image")
ax4.set_title("Scattering Curve")
draw_voxles_as_dots(ax1, canvas)
# relation 3: Fourier transform
F = np.fft.fftn(canvas)
# relation 2: reciprocal space image
ft_image = np.abs(F)
draw_voxles_as_dots(ax2, ft_image**2)
# relation 2 and 1: detector image and scattering curve
# the spherical average and circular average is done in get_detector_info
q = np.linspace(0.005, 0.5, 100)
info = get_detector_info(q, F)
# here, detector image is generated from the scattering curve
# for illustration purpose which is the reverse of relation 1
draw_detector_image(ax3, q, info.y)
ax4.set_xlabel("q")
ax4.set_ylabel("I(q)")
ax4.plot(q, info.y)
ax1.set_xlim(ax2.get_xlim())
ax1.set_ylim(ax2.get_ylim())
ax1.set_zlim(ax2.get_zlim())
fig.tight_layout()
plot_an_ellipsoid_with_ft_squared_and_detector_image(32, (16,16,16), 5, 4, 2)
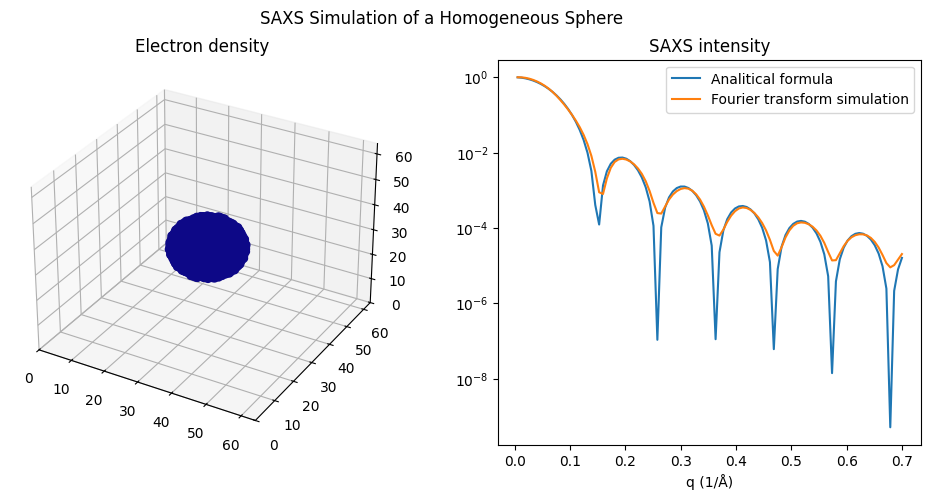
We can confirm the above relationships by computing the scattering curve in two ways: using the analytical formula and by direct Fourier transform, as shown below.
from molass import get_version
assert get_version() >= '0.6.0', "This tutorial requires molass version 0.6.0 or higher."
import numpy as np
import matplotlib.pyplot as plt
from molass.Shapes import Sphere
from molass.DensitySpace import VoxelSpace
from molass.SAXS.Simulator import compute_saxs
from molass.SAXS.Models.Formfactors import homogeneous_sphere
# %matplotlib widget
q = np.linspace(0.005, 0.7, 100)
R = 10
I = homogeneous_sphere(q, 3*R) # why 3*R?
sphere = Sphere(radius=10)
space = VoxelSpace(64, sphere)
saxs = compute_saxs(space.rho, q=q, dmax=64)
fig = plt.figure(figsize=(12, 5))
fig.suptitle('SAXS Simulation of a Homogeneous Sphere')
ax1 = fig.add_subplot(121, projection='3d')
ax1.set_title('Electron density')
space.plot_as_dots(ax1)
ax2 = fig.add_subplot(122)
ax2.set_title('SAXS intensity')
ax2.set_yscale('log')
ax2.plot(q, I, label='Analitical formula')
curve = saxs.get_curve()
ax2.plot(q, curve.y, label='Fourier transform simulation')
ax2.legend()
ax2.set_xlabel('q (1/Å)')
Text(0.5, 0, 'q (1/Å)')
from molass.Shapes import Ellipsoid
from molass.DensitySpace import VoxelSpace
from molass.SAXS.Simulator import compute_saxs
from molass.SAXS.Models.Formfactors import tri_axial_ellipsoid
def plot_tri_axial_ellipsoid(a, b, c):
I = tri_axial_ellipsoid(q, 3*a, 3*b, 3*c) # why 3*a, 3*b, 3*c?
ellipsoid = Ellipsoid(a, b, c)
space = VoxelSpace(64, ellipsoid)
saxs = compute_saxs(space.rho, q=q, dmax=64)
fig = plt.figure(figsize=(12, 5))
fig.suptitle('SAXS Simulation of a Tri-axial Ellipsoid')
ax1 = fig.add_subplot(121, projection='3d')
ax1.set_title('Electron density')
space.plot_as_dots(ax1)
ax2 = fig.add_subplot(122)
ax2.set_title('SAXS intensity')
ax2.set_yscale('log')
ax2.plot(q, I, label='Tri-axial Ellipsoid')
curve = saxs.get_curve()
ax2.plot(q, curve.y, label='Fourier transform simulation')
ax2.legend()
ax2.set_xlabel('q (1/Å)')
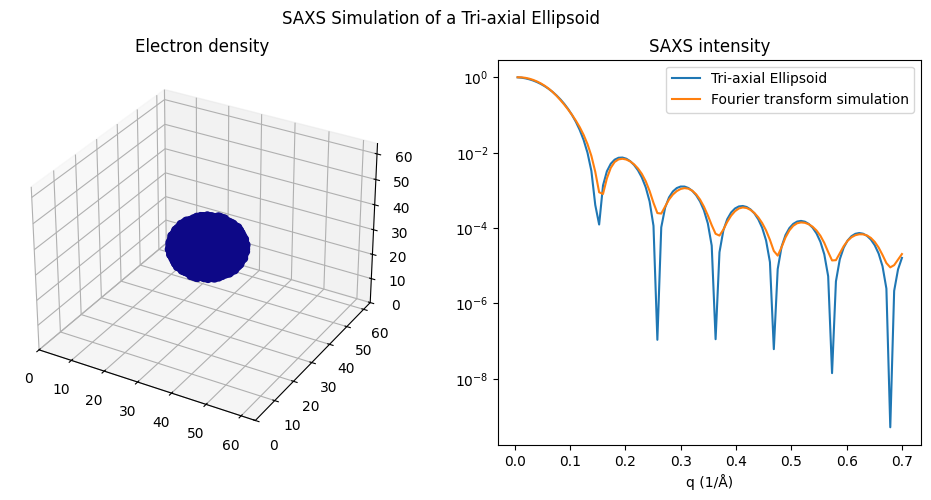
With the current state of the code, the calculation for an ellipsoid may be inaccurate (see the next plot), while it appears correct when the semi-axes are equal (i.e., \( a=b=c \), as in the followed plot resulting in a sphere).
Note
The code is for illustration purposes only and is not used in real analysis.
plot_tri_axial_ellipsoid(10, 8, 6)
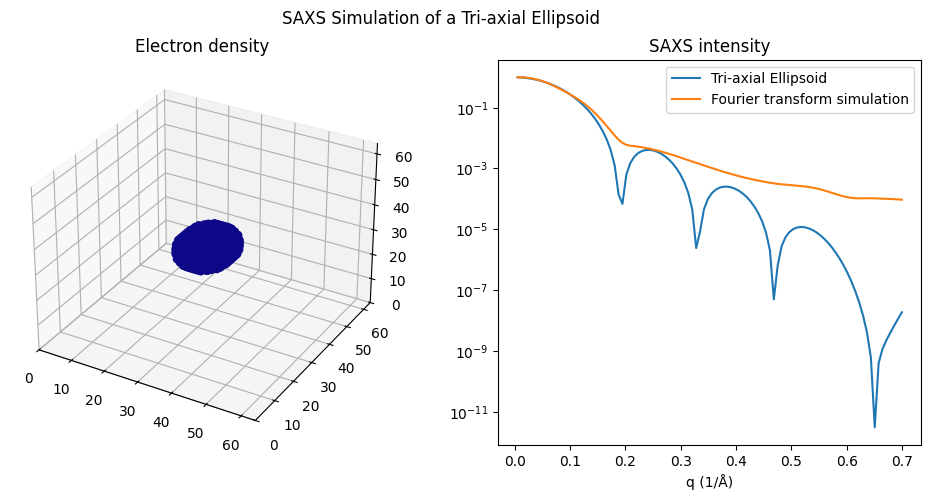
plot_tri_axial_ellipsoid(10, 10, 10)