3.1. Matrix Multiplication#
In this section, we will confirm that the data from SEC-SAXS experiments can be represented as matrices which are products of the curves. This fact is expressed in a formula as follows.
where
\( M \) : measured data
\( P \) : spectral curves
\( C \) : elutional curves
\( P \cdot C \) : matrix multiplication
Note
We refer to the curves related to elution as ‘elutional curves’ for contrast with ‘spectral curves.
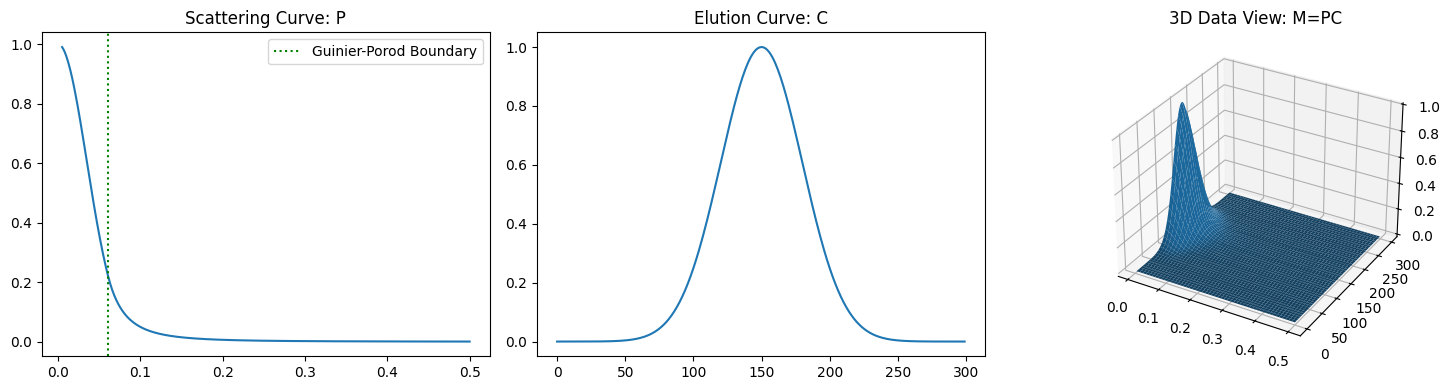
3.1.1. Case of Single Component#
For a case with a single component, confirm the following points.
The \(P\) matrix consists of a single column vector which represents the spectral curve.
The \(C\) matrix consists of a single row vector which represents the eluional curve.
import numpy as np
import matplotlib.pyplot as plt
from molass.SAXS.Models.Simple import guinier_porod
from molass.SEC.Models.Simple import gaussian
x = np.arange(300)
q = np.linspace(0.005, 0.5, 400)
def plot_single_component_data(scatter_params, elution_params):
G, Rg, d = scatter_params
h, m, s = elution_params
fig = plt.figure(figsize=(15,4))
ax1 = fig.add_subplot(131)
ax2 = fig.add_subplot(132)
ax3 = fig.add_subplot(133, projection='3d')
I, q1 = guinier_porod(q, G, Rg, d, return_also_q1=True)
# ax1.set_yscale('log')
ax1.set_title("Scattering Curve: P")
ax1.plot(q, I)
ax1.axvline(q1, linestyle=':', color="green", label='Guinier-Porod Boundary')
ax1.legend()
y = gaussian(x, h, m, s)
ax2.set_title("Elution Curve: C")
ax2.plot(x, y)
P = I.reshape((len(q),1)) # make it a matrix
C = y.reshape((1,len(x))) # make it a matrix
M = P @ C # matrix multiplication
xx, qq = np.meshgrid(x, q)
ax3.set_title("3D Data View: M=PC")
ax3.plot_surface(qq, xx, M)
fig.tight_layout()
plot_single_component_data((1, 35, 3), (1, 150, 30))
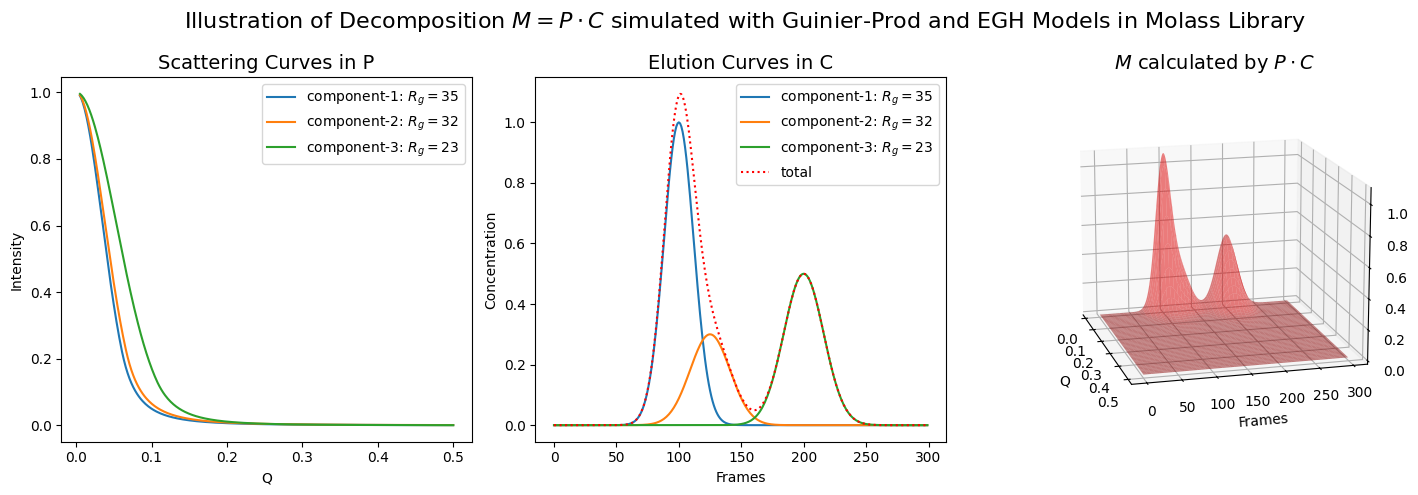
3.1.2. Case of Multiple Components#
For a case with multiple components, confirm the following points.
The \(P\) matrix consists of multiple column vectors which represent the spectral curves.
The \(C\) matrix consists of multiple row vectors which represent the eluional curves.
def plot_multiple_component_data(scattering_params, elution_params, use_matrices=False, view=None):
fig = plt.figure(figsize=(15,5))
ax1 = fig.add_subplot(131)
ax2 = fig.add_subplot(132)
ax3 = fig.add_subplot(133, projection='3d')
fig.suptitle(r"Illustration of Decomposition $M = P \cdot C$ simulated with Guinier-Prod and EGH Models in Molass Library", fontsize=16)
# ax1.set_yscale('log')
ax1.set_title("Scattering Curves in P", fontsize=14)
ax1.set_xlabel("Q")
ax1.set_ylabel("Intensity")
w_list = []
rgs = []
for i, (G, Rg, d) in enumerate(scattering_params):
I, q1 = guinier_porod(q, G, Rg, d, return_also_q1=True)
w_list.append(I)
color = "C%d" % i
ax1.plot(q, I, color=color, label='component-%d: $R_g=%.3g$' % (i+1, Rg))
rgs.append(Rg)
# ax1.axvline(q1, linestyle=':', color=color, label='Guinier-Porod $Q_1$')
ax1.legend()
y_list = []
ax2.set_title("Elution Curves in C", fontsize=14)
ax2.set_xlabel("Frames")
ax2.set_ylabel("Concentration")
for i, (h, m, s) in enumerate(elution_params):
y = gaussian(x, h, m, s)
y_list.append(y)
ax2.plot(x, y, label='component-%d: $R_g=%.3g$' % (i+1, rgs[i]))
ty = np.sum(y_list, axis=0)
ax2.plot(x, ty, ':', color='red', label='total')
ax2.legend()
ax3.set_title(r"$M$ calculated by $P \cdot C$", fontsize=14)
ax3.set_xlabel("Q")
ax3.set_ylabel("Frames")
ax3.set_zlabel("Intensity")
xx, qq = np.meshgrid(x, q)
if use_matrices:
P = np.array(w_list).T
C = np.array(y_list)
M = P @ C
else:
zz_list = []
for w, y in zip(w_list, y_list):
zz = w.reshape((len(q),1)) @ y.reshape((1,len(x)))
zz_list.append(zz)
M = np.sum(zz_list, axis=0)
ax3.plot_surface(qq, xx, M, color='red', alpha=0.5)
# ax3.legend()
if view is not None:
ax3.view_init(azim=view[0], elev=view[1])
fig.tight_layout()
fig.subplots_adjust(right=0.95)
rgs = (35, 32, 23)
scattering_params = [(1, rgs[0], 3), (1, rgs[1], 3), (1, rgs[2], 4)]
elution_params = [(1, 100, 12), (0.3, 125, 16), (0.5, 200, 16)]
plot_multiple_component_data(scattering_params, elution_params, use_matrices=True, view=(-15, 15))