9. Nontrivial Decomposition#
This chapter will discuss the cases where component peaks are not apparent.
9.1. Initial Observation#
Let us first observe such an example.
from molass import get_version
assert get_version() >= '0.6.0', "This tutorial requires molass version 0.6.0 or higher."
from molass_data import get_version
assert get_version() >= '0.3.0', "This tutorial requires molass_data version 0.3.0 or higher."
from molass_data import SAMPLE4
from molass.DataObjects import SecSaxsData as SSD
ssd = SSD(SAMPLE4)
trimmed_ssd = ssd.trimmed_copy()
corrected_ssd = trimmed_ssd.corrected_copy()
corrected_ssd.plot_compact();
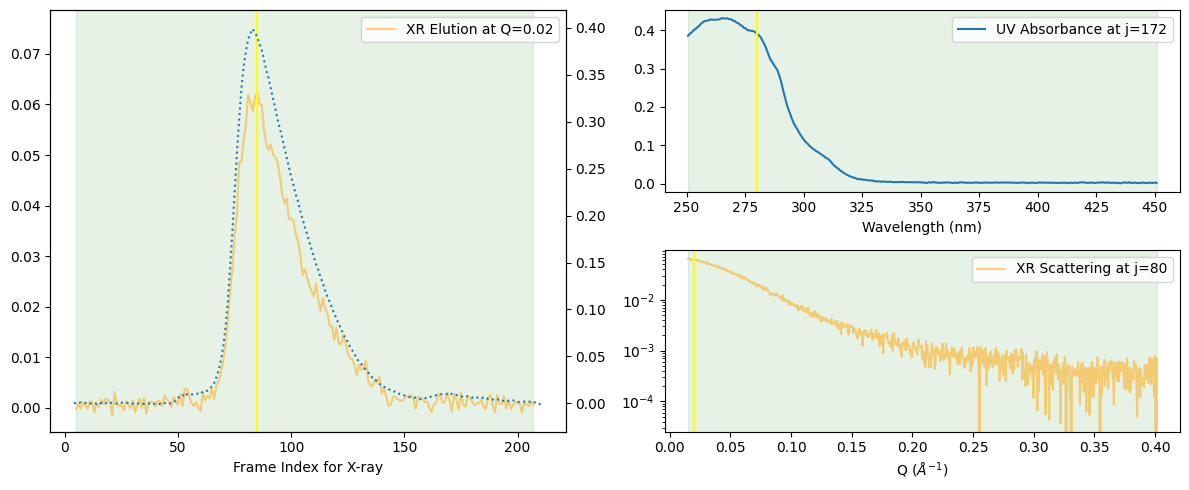
At first glance, the peak may seem to consist of a single component. For a more detailed observation, let us assume it may consist of two components and add the Rg curve.
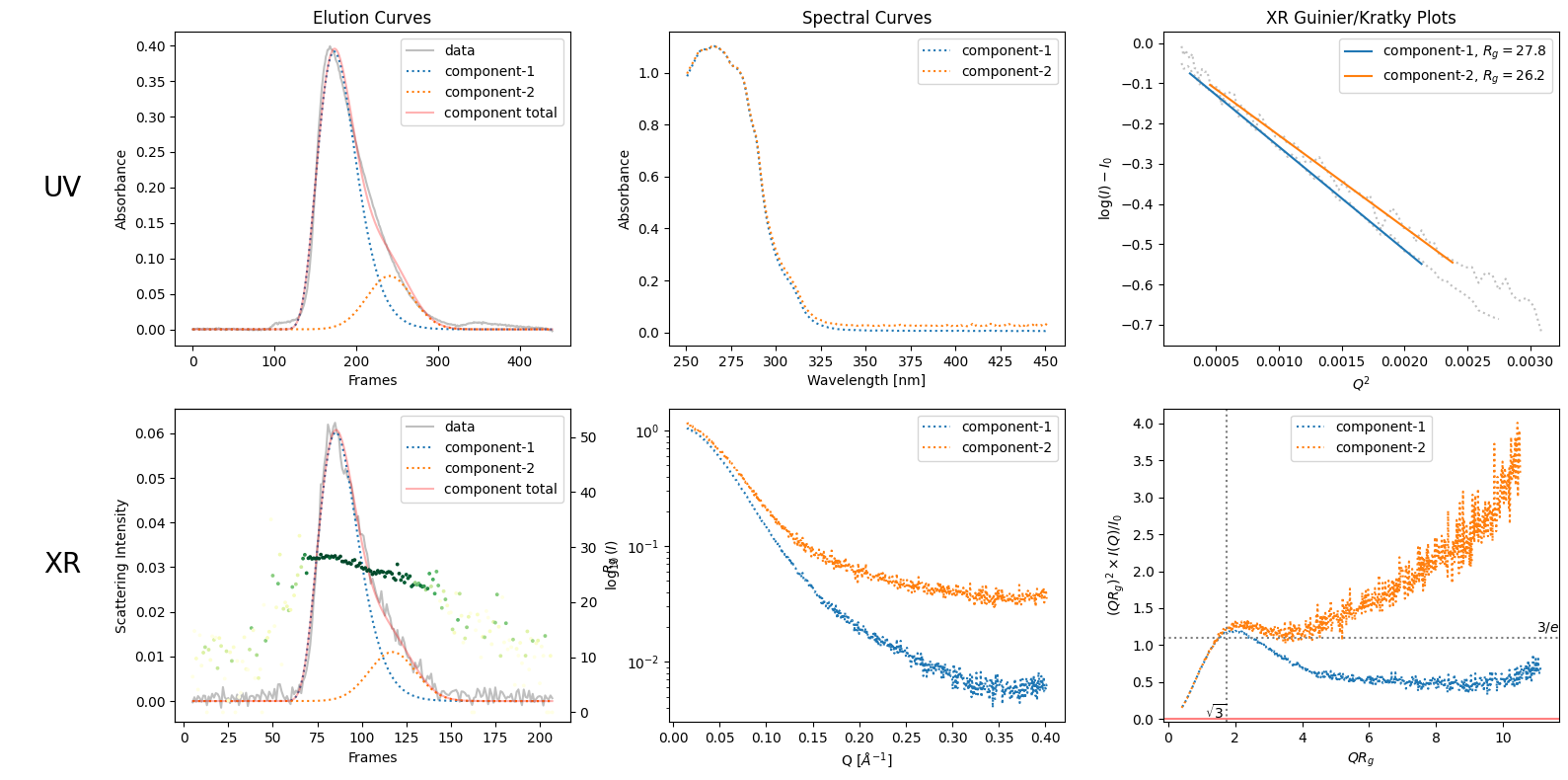
rgcurve = corrected_ssd.xr.compute_rgcurve()
decomposition = corrected_ssd.quick_decomposition(num_components=2)
decomposition.plot_components(rgcurve=rgcurve)
100%|██████████| 203/203 [00:07<00:00, 25.46it/s]
<molass.PlotUtils.PlotResult.PlotResult at 0x24a368a6f60>
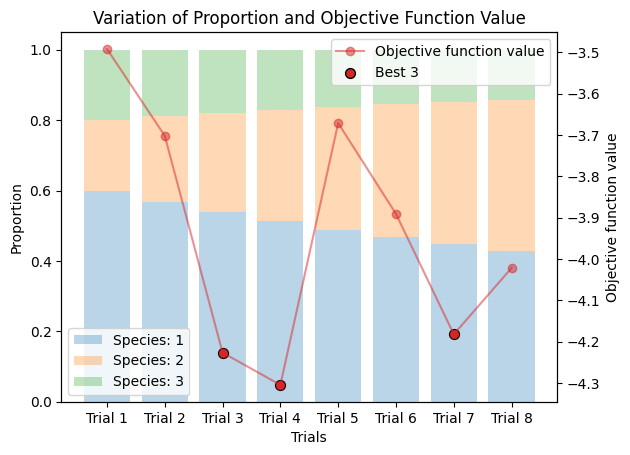
9.2. Varied Binary Proportions#
If you are not sure about the default compositiun, you can compare the results with different proportions as follows.
import numpy as np
num_trails = 8
species1_proportions = np.ones(num_trails) * 3
species2_proportions = np.linspace(1, 3, num_trails)
proportions = np.array([species1_proportions, species2_proportions]).T
proportions
array([[3. , 1. ],
[3. , 1.28571429],
[3. , 1.57142857],
[3. , 1.85714286],
[3. , 2.14285714],
[3. , 2.42857143],
[3. , 2.71428571],
[3. , 3. ]])
corrected_ssd.plot_varied_decompositions(proportions, rgcurve=rgcurve, best=3)
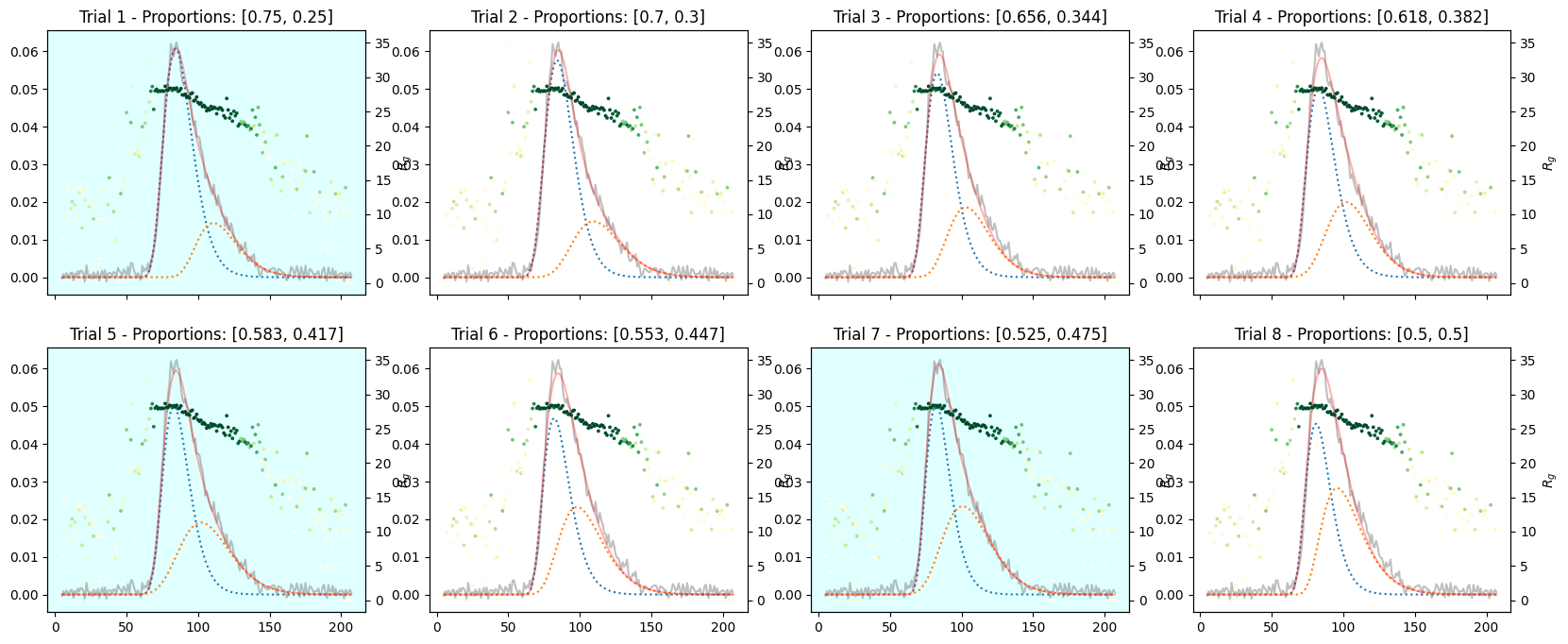
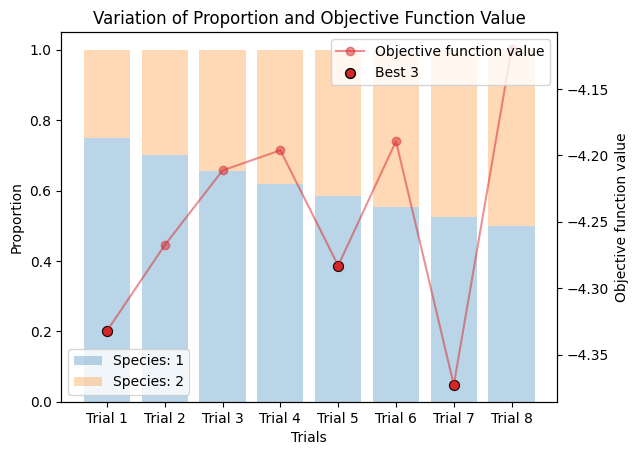
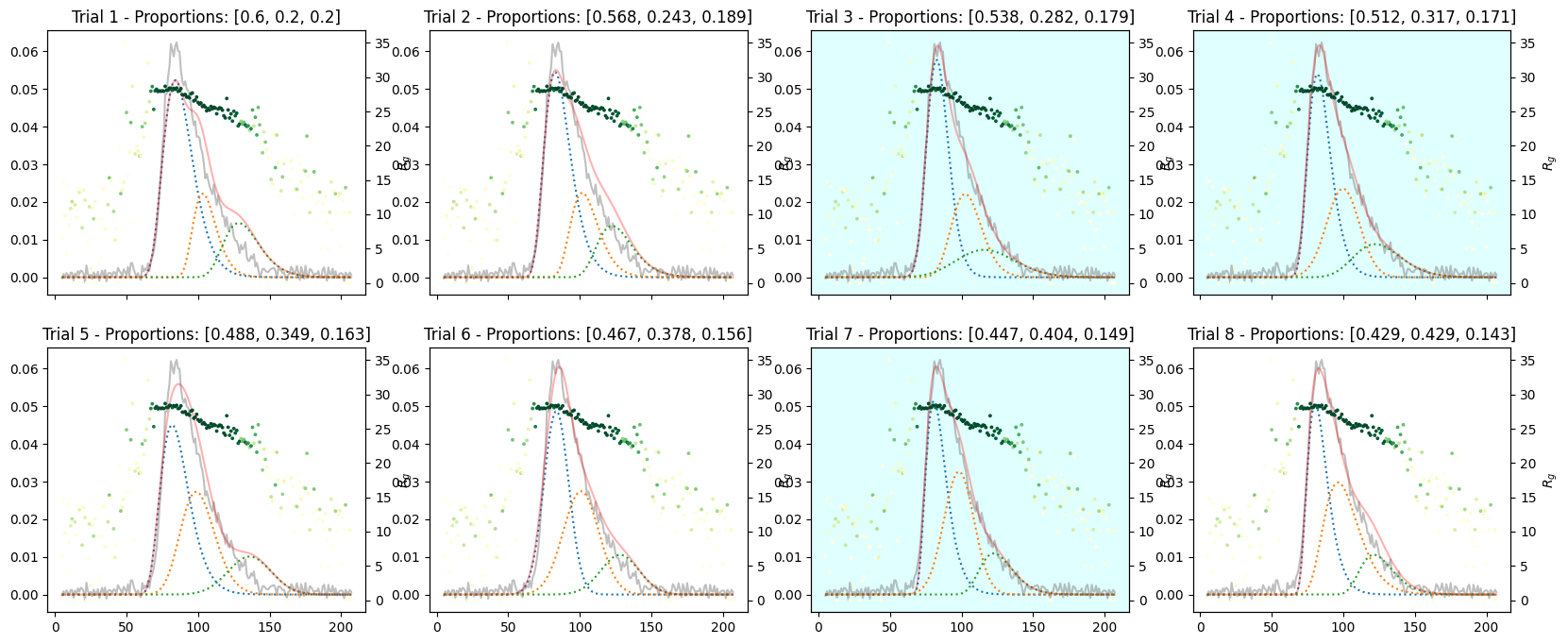
9.3. Varied Tertiary Proportions#
If the existence of three components are suspected, do as follows.
species3_proportions = np.ones(num_trails) * 1
proportions = np.array([species1_proportions, species2_proportions, species3_proportions]).T
proportions
array([[3. , 1. , 1. ],
[3. , 1.28571429, 1. ],
[3. , 1.57142857, 1. ],
[3. , 1.85714286, 1. ],
[3. , 2.14285714, 1. ],
[3. , 2.42857143, 1. ],
[3. , 2.71428571, 1. ],
[3. , 3. , 1. ]])
corrected_ssd.plot_varied_decompositions(proportions, rgcurve=rgcurve, best=3)