7.2. Proportional Constaints#
from molass import get_version
assert get_version() >= '0.5.2', "This tutorial requires molass version 0.5.2 or higher."
from molass_data import get_version
assert get_version() >= '0.3.0', "This tutorial requires molass_data version 0.3.0 or higher."
from molass_data import SAMPLE4
from molass.DataObjects import SecSaxsData as SSD
ssd = SSD(SAMPLE4)
trimmed_ssd = ssd.trimmed_copy()
corrected_ssd = trimmed_ssd.corrected_copy()
import numpy as np
import matplotlib.pyplot as plt
from molass.Decompose.Proportional import decompose_proportionally
icurve = corrected_ssd.xr.get_icurve()
x, y = icurve.get_xy()
result = decompose_proportionally(x, y, [3,2,1], debug=True)
[0.71855534 0.10826109 0.17318356]
[0.49999669 0.33334145 0.16666185]
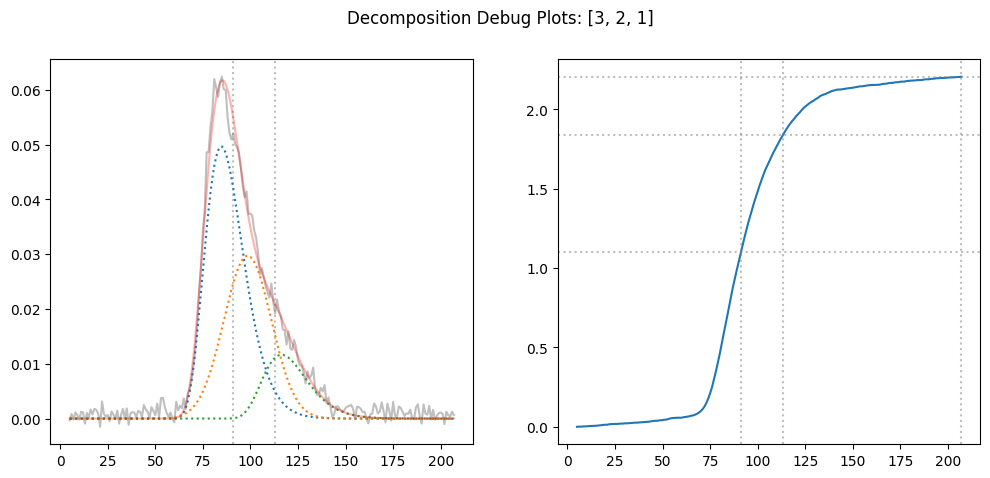
result
message: Maximum number of function evaluations has been exceeded.
success: False
status: 1
fun: -4.262727985617209
x: [ 5.122e-02 8.156e+01 7.883e+00 5.251e+00 2.585e-02
9.890e+01 1.083e+01 -6.481e-03 1.128e-02 1.204e+02
1.206e+01 6.805e+00]
nit: 1627
nfev: 2400
final_simplex: (array([[ 5.122e-02, 8.156e+01, ..., 1.206e+01,
6.805e+00],
[ 5.122e-02, 8.156e+01, ..., 1.206e+01,
6.805e+00],
...,
[ 5.122e-02, 8.156e+01, ..., 1.206e+01,
6.805e+00],
[ 5.122e-02, 8.156e+01, ..., 1.206e+01,
6.805e+00]]), array([-4.263e+00, -4.263e+00, ..., -4.263e+00, -4.263e+00]))
len(x)
203
rgcurve = corrected_ssd.xr.compute_rgcurve()
100%|██████████| 203/203 [00:04<00:00, 45.35it/s]
num_trails = 8
species1_proportions = np.ones(num_trails) * 3
species2_proportions = np.linspace(1, 3, num_trails)
proportions = np.array([species1_proportions, species2_proportions]).T
proportions
array([[3. , 1. ],
[3. , 1.28571429],
[3. , 1.57142857],
[3. , 1.85714286],
[3. , 2.14285714],
[3. , 2.42857143],
[3. , 2.71428571],
[3. , 3. ]])
from importlib import reload
import molass.Decompose.VaryUtils
reload(molass.Decompose.VaryUtils)
from molass.Decompose.VaryUtils import plot_varied_decompositions
plot_varied_decompositions(x, y, proportions, rgcurve=rgcurve, best=3)
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[7], line 4
2 import molass.Decompose.VaryUtils
3 reload(molass.Decompose.VaryUtils)
----> 4 from molass.Decompose.VaryUtils import plot_varied_decompositions
5 plot_varied_decompositions(x, y, proportions, rgcurve=rgcurve, best=3)
ImportError: cannot import name 'plot_varied_decompositions' from 'molass.Decompose.VaryUtils' (c:\Program Files\Python312\Lib\site-packages\molass\Decompose\VaryUtils.py)
species3_proportions = np.ones(num_trails) * 1
proportions = np.array([species1_proportions, species2_proportions, species3_proportions]).T
proportions
plot_varied_decompositions(x, y, proportions, rgcurve=rgcurve, best=3)
from molass_data import SAMPLE1
ssd = SSD(SAMPLE1)
trimmed_ssd = ssd.trimmed_copy()
corrected_ssd = trimmed_ssd.corrected_copy()
modified_decomposition = corrected_ssd.quick_decomposition(num_components=3, proportions=[0.32, 0.20, 0.48])
plot2 = modified_decomposition.plot_components(title="Modified Decomposition of Sample1 (num_components=3, proportions=[0.32, 0.20, 0.48])")
